| Structure-Driven Unsupervised Domain Adaptation for Cross-Modality Cardiac Segmentation | |||
| Zhiming Cui1,2, Changjian Li3, Zhixu Du1, Nenglun Chen1, Guodong Wei1, Runnan Chen1, Lei Yang1, Dinggang Shen2, Wenping Wang1,5 |
1The University of Hong Kong, 2 ShanghaiTech University, 3 University College London, 4Texas A&M University |
||
| IEEE Transactions on Medical Imaging (TMI), 2021 |
|||
 |
|||
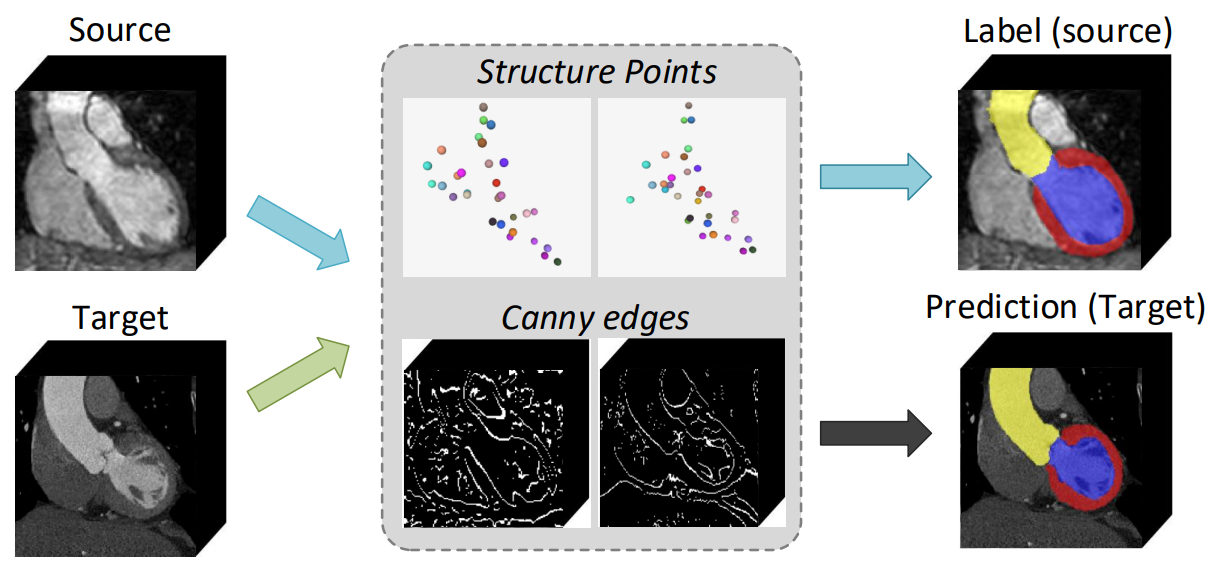
| Fig. 1. Overview of our unsupervised domain adaptation method on cardiac MRI and CT images. The example shows MRI (source) to CT (target) adaptation. Specifically, our method aims to extract the cardiac structure across modalities by a set of landmarks, while accurately predict object labels in the target domain guided by the domain-invariant structural landmarks and edge information. | |||
| Abstract | |||
| Performance degradation due to domain shift remains a major challenge in medical image analysis. Unsupervised domain adaptation that transfers knowledge learned from the source domain with ground truth labels to the target domain without any annotation is the mainstream solution to resolve this issue. In this paper, we present a novel unsupervised domain adaptation framework for cross-modality cardiac segmentation, by explicitly capturing a common cardiac structure embedded across different modalities to guide cardiac segmentation. In particular, we first extract a set of 3D landmarks, in a selfsupervised manner, to represent the cardiac structure of different modalities. The high-level structure information is then combined with another complementary feature, the Canny edges, to produce accurate cardiac segmentation results both in the source and target domains. We extensively evaluate our method on the MICCAI 2017 MM-WHS dataset for cardiac segmentation. The evaluation, comparison and comprehensive ablation studies demonstrate that our approach achieves satisfactory segmentation results and outperforms state-of-the-art unsupervised domain adaptation methods by a significant margin. | |||
|
|||
|
|
|||
| Network Overview | |||
 |
|||
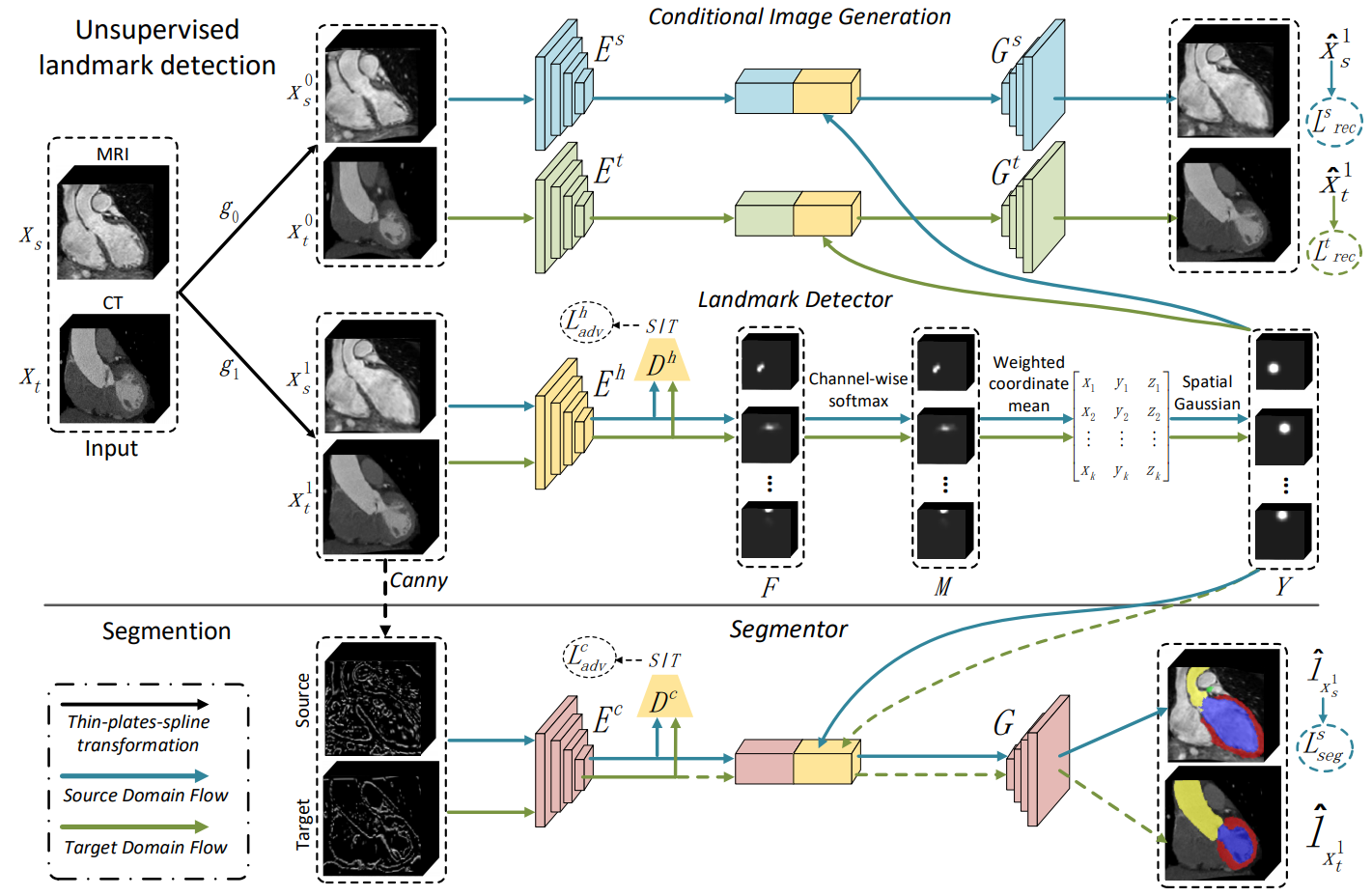
| Fig. 2. The framework of our method for unsupervised domain adaptation. The landmark detection module first extracts the anatomical cardiac structure represented by a set of 3D landmarks. The disentangle process is achieved by the conditional image generation mechanism. Then, the segmentation module takes as input both the extracted structure and a set of edges from the Canny operator to produce reliable results. | |||
| Results | |||
| We have evaluated our method on the public dataset, compared it against state-of-the-art methods and conducted comprehensive ablation studies to validate the effectiveness and accuracy. All experiments are performed on a machine with an Intel(R) Xeon(R) V4 1.9GHz CPU, 4 Nvidia 1080Ti GPUs, and 32GB. Below are some representative results, more results and inspiring discussions can be found in the paper. | |||
| Result Gallery | |||
 |
|||
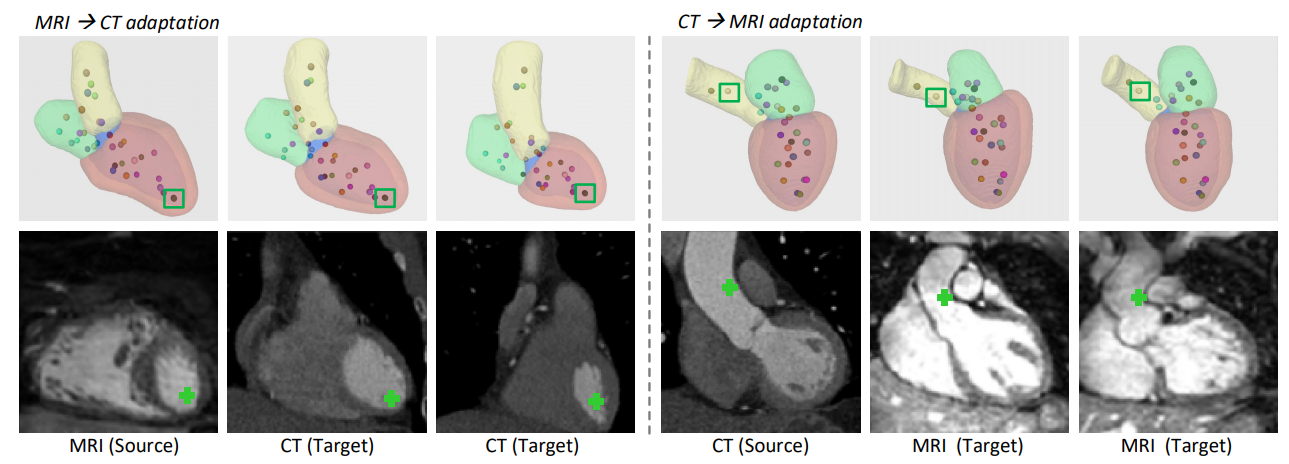
| Fig. 4. 3D segmentation results and corresponding extracted landmarks, including MRI to CT adaptation (left) and CT to MRI adaptation (right). The first row overlays the segmented cardiac sub-structures with spatial landmarks, where the color-coding of landmarks expresses the consistency across modalities. The second row visualizes the 2D coronal view image slice on the location of the selected landmark point (green box). Each column corresponds to one example. | |||
| Comparison | |||
 |
|||
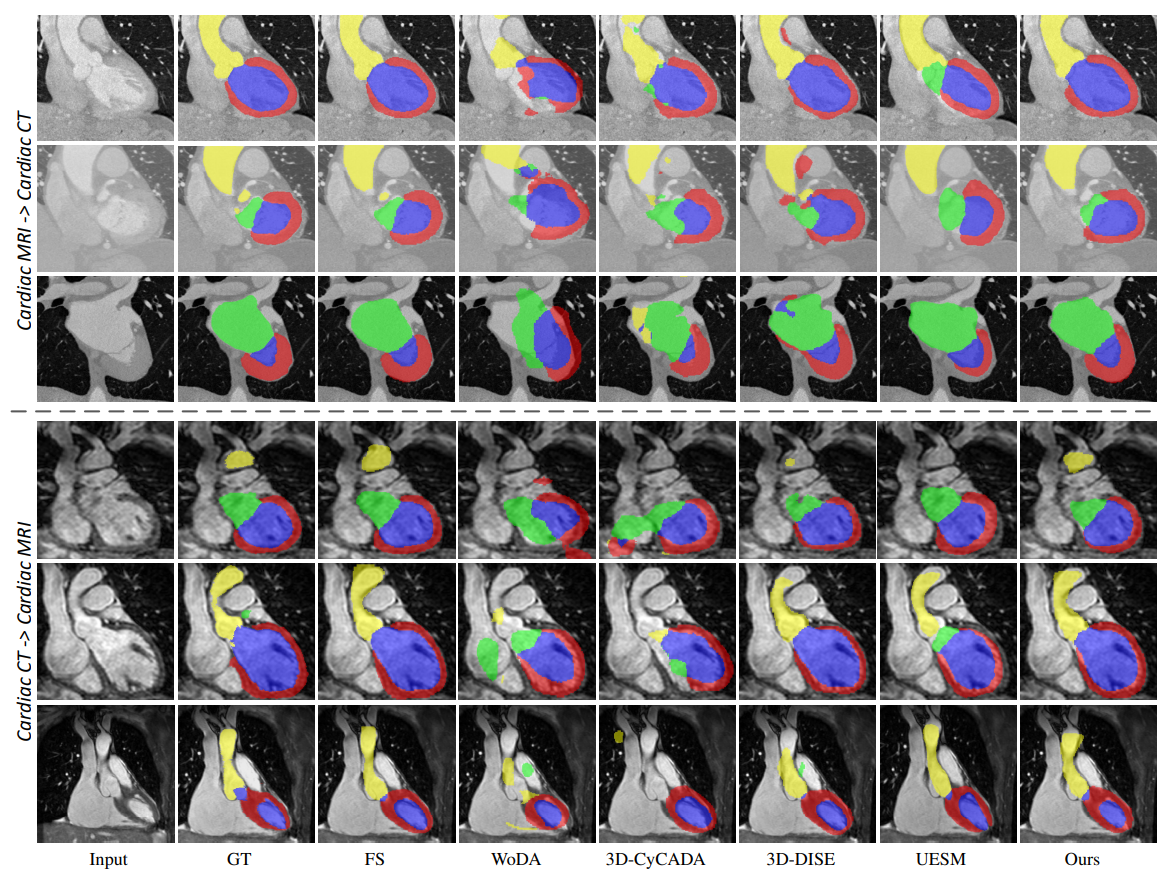
| Fig. 5. Qualitative results of different methods for unsupervised MRI to CT domain adaptation (top three rows) and CT to MRI domain adaptation (bottom three rows). Typical examples are showed row-by-row. The yellow, green, red and blue colors represent the cardiac structures AA, LA-BC, LV-BC and MYO, respectively. | |||
|
|
|||
| ©Changjian Li. Last update: June, 2021. |